Maternally supplied mRNAs and proteins, produced by over half of the coding genes in zebrafish, control oogenesis and the earliest processes of embryonic development, especially those before zygotic genome activation. Some maternal factors can also perform zygotic functions during early development. Thus, eliminating both maternal and zygotic products is essential to elucidate functional roles of genes with maternal expression. However, most maternal gene products are cell-autonomously deposited in primary oocytes that have the same genotype as somatic cells. As a result, to generate maternal (M) or maternal-zygotic (MZ) mutant offspring by traditional genetic approach, viable and fertile homozygous mutant females are indispensable. These homozygous mutants are classically produced in a time-consuming manner by repeated crossing and screening through typically three generations (almost nine months in zebrafish). Nevertheless, if loss of zygotic gene functions causes death or sterility, it becomes particularly difficult to produce maternal mutants. One way to circumvent these obstacles is to rescue phenotype defects by injecting the wild-type mRNA, such as the case for the generation of Moep mutants, but this approach was proved to be possible only for a small proportion of zygotic lethal genes. Several alternatives have also been developed but these methods remain technically challenging, time-consuming, or less efficient. Therefore, a rapid and straightforward system to inactivate genes in the oocyte may break the barrier to study their maternal functions.

Prof. Shao’s group developed a rapid conditional knockout strategy to generate maternal mutants by just one generation (less than three months). By introducing a single or multiple sgRNA expression modules into Tg(zpc:zcas9) embryos via I-SceI-mediated transgenesis, they successfully generated maternal mutants for nanog and ctnnb2 (β-catenin2) genes among F1 embryos (Figure 1). They also discovered a dramatic tendency for genome editing in zebrafish oocytes to frequently produce heritable large deletions when expressing multiple sgRNAs (Figure 2). Therefore, their study demonstrates the promising potential of this novel conditional knockout strategy to circumvent current technical restrictions in the functional studies of maternal factors.
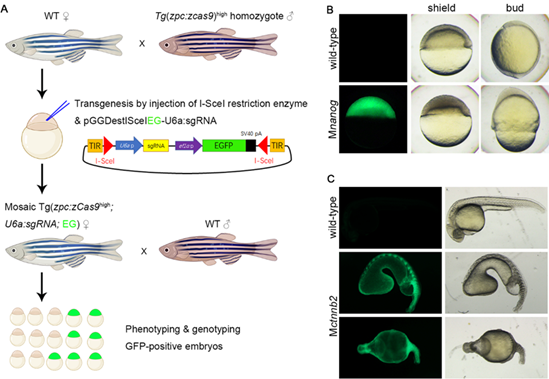
Figure1, Procedure of rapid oocyte-specific conditional knockout
A, workflow to obtain maternal mutants through oocyte-specific conditional genome editing.
B, Using this method we obtained nanog maternal mutants, which showed extremely epiboly delay.
C, The constructed ctnnb2 maternal mutants displayed severe defects in body axis formation.

Figure2, Expression of Cas9 and multiple sgRNAs in oocytes tends to generate large deletions in nanog gene.
This work was published in the Science Advances on August 6th. Prof. Ming Shao at School of Life Sciences, Shandong University was the corresponding author of this paper. This work was supported by the National Natural Science Foundation of China (grant numbers 31871451 and 32070813), the National Key R&D Program of China (grant number 2018YFA0801000), and the Program of Shandong University Qilu Young Scholars.
Paper link:
https://advances.sciencemag.org/content/7/32/eabg4243